RBF核的顯式特征映射近似簡介
本文用一個示例介紹了近似RBF核的特征映射的方法。
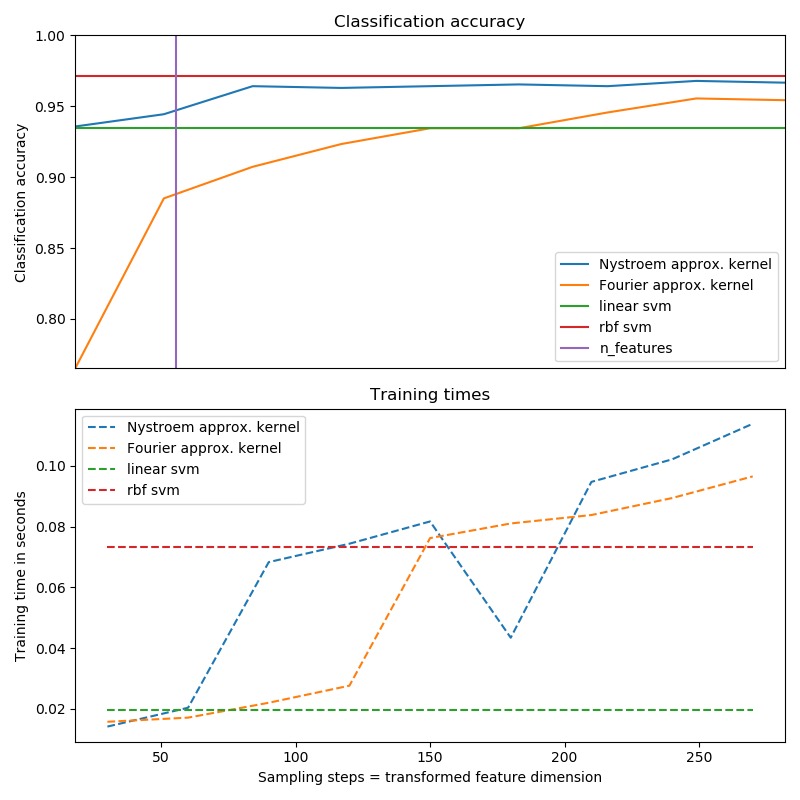
具體來說,示例中展示了在便用SVM對數字數據集進行分類的情況下,如何使用RBFSampler和Nystroem來近似RBF核的特征映射。其中比較了使用原始空間中的線性SVM,使用近似映射和使用內核化SVM的結果。不同模型運行時間和精度的比較涉及:不同蒙特卡洛采樣樣本數量(對於RBFSampler,它使用隨機傅立葉特征)和訓練集的不同大小子集(用於Nystroem)。
請注意,核近似的主要優勢在於性能提升,但這裏的數據集規模不足以顯示核近似的好處,因為精確的SVM仍然相當快。
對更多維度進行采樣顯然會帶來更好的分類結果,但代價更高。這意味著在運行時間和精度之間需要權衡,這由參數n_components給出。請注意,通過使用隨機梯度下降法(sklearn.linear_model.SGDClassifier)可以大大加快求解線性SVM以及近似核SVM的速度。對於有核函數的SVM,這是不容易實現的。
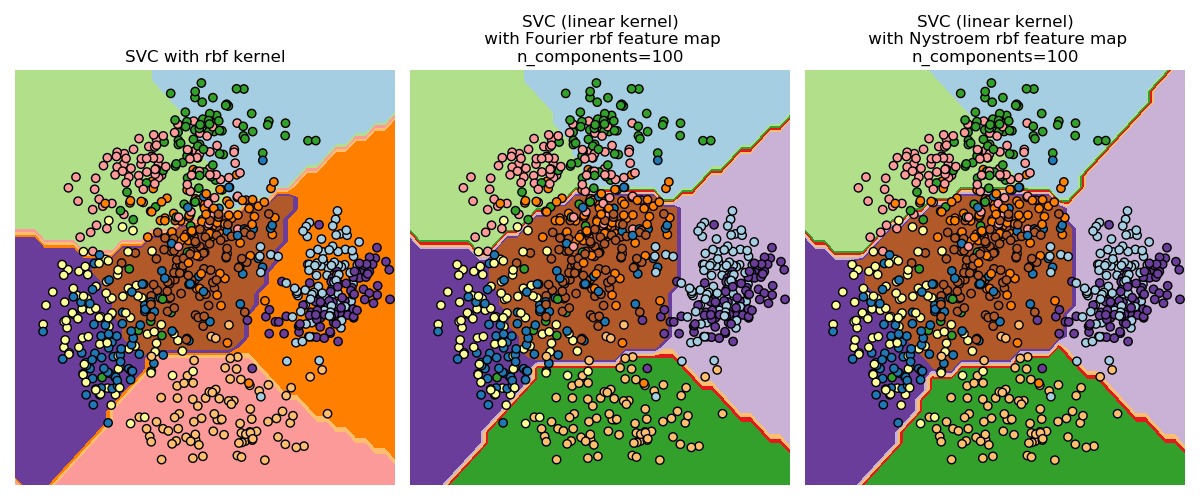
第二個圖顯示了RBF核SVM和帶有近似核映射的線性SVM的決策麵。該圖顯示了投影到數據的前兩個主要成分上的分類器的決策麵。注意,這種可視化結果是否完全準確是存疑的,因為它隻是決策麵上64個維度中的一個有趣切片。特別要注意的是,數據點(用點表示)不一定要分類到它所在的區域,因為它不會位於前兩個主要成分所跨越的平麵上。
對於RBFSampler和Nystroem的詳細用法,請參考文檔核近似。
代碼實現[Python]
# -*- coding: utf-8 -*-
print(__doc__)
# Author: Gael Varoquaux
# Andreas Mueller
# License: BSD 3 clause
# 導入Python標準科學計算相關模塊
import matplotlib.pyplot as plt
import numpy as np
from time import time
# 導入數據集、分類器、性能評估標準
from sklearn import datasets, svm, pipeline
from sklearn.kernel_approximation import (RBFSampler,
Nystroem)
from sklearn.decomposition import PCA
# 手寫數字數據集
digits = datasets.load_digits(n_class=9)
# 為了在數據集上應用分類器,我們需要展平圖像到一個維度,將樣本數據集轉為(samples, feature)矩陣的形式:
n_samples = len(digits.data)
data = digits.data / 16.
data -= data.mean(axis=0)
# 使用1/2的數字數據集作為訓練集
data_train, targets_train = (data[:n_samples // 2],
digits.target[:n_samples // 2])
# 另外1/2作為測試集
data_test, targets_test = (data[n_samples // 2:],
digits.target[n_samples // 2:])
# data_test = scaler.transform(data_test)
# 創建SVM分類器
kernel_svm = svm.SVC(gamma=.2)
linear_svm = svm.LinearSVC()
# 創建核近似pipeline,並添加線性SVM
feature_map_fourier = RBFSampler(gamma=.2, random_state=1)
feature_map_nystroem = Nystroem(gamma=.2, random_state=1)
fourier_approx_svm = pipeline.Pipeline([("feature_map", feature_map_fourier),
("svm", svm.LinearSVC())])
nystroem_approx_svm = pipeline.Pipeline([("feature_map", feature_map_nystroem),
("svm", svm.LinearSVC())])
# 使用線性SVM和核SVM做擬合和預測
kernel_svm_time = time()
kernel_svm.fit(data_train, targets_train)
kernel_svm_score = kernel_svm.score(data_test, targets_test)
kernel_svm_time = time() - kernel_svm_time
linear_svm_time = time()
linear_svm.fit(data_train, targets_train)
linear_svm_score = linear_svm.score(data_test, targets_test)
linear_svm_time = time() - linear_svm_time
sample_sizes = 30 * np.arange(1, 10)
fourier_scores = []
nystroem_scores = []
fourier_times = []
nystroem_times = []
for D in sample_sizes:
fourier_approx_svm.set_params(feature_map__n_components=D)
nystroem_approx_svm.set_params(feature_map__n_components=D)
start = time()
nystroem_approx_svm.fit(data_train, targets_train)
nystroem_times.append(time() - start)
start = time()
fourier_approx_svm.fit(data_train, targets_train)
fourier_times.append(time() - start)
fourier_score = fourier_approx_svm.score(data_test, targets_test)
nystroem_score = nystroem_approx_svm.score(data_test, targets_test)
nystroem_scores.append(nystroem_score)
fourier_scores.append(fourier_score)
# 繪製結果圖:
plt.figure(figsize=(8, 8))
accuracy = plt.subplot(211)
# second y axis for timeings
timescale = plt.subplot(212)
accuracy.plot(sample_sizes, nystroem_scores, label="Nystroem approx. kernel")
timescale.plot(sample_sizes, nystroem_times, '--',
label='Nystroem approx. kernel')
accuracy.plot(sample_sizes, fourier_scores, label="Fourier approx. kernel")
timescale.plot(sample_sizes, fourier_times, '--',
label='Fourier approx. kernel')
# 水平線用於精確的rbf和線性內核
accuracy.plot([sample_sizes[0], sample_sizes[-1]],
[linear_svm_score, linear_svm_score], label="linear svm")
timescale.plot([sample_sizes[0], sample_sizes[-1]],
[linear_svm_time, linear_svm_time], '--', label='linear svm')
accuracy.plot([sample_sizes[0], sample_sizes[-1]],
[kernel_svm_score, kernel_svm_score], label="rbf svm")
timescale.plot([sample_sizes[0], sample_sizes[-1]],
[kernel_svm_time, kernel_svm_time], '--', label='rbf svm')
# 垂直線用於數據集維度 = 64
accuracy.plot([64, 64], [0.7, 1], label="n_features")
# legends and labels
accuracy.set_title("Classification accuracy")
timescale.set_title("Training times")
accuracy.set_xlim(sample_sizes[0], sample_sizes[-1])
accuracy.set_xticks(())
accuracy.set_ylim(np.min(fourier_scores), 1)
timescale.set_xlabel("Sampling steps = transformed feature dimension")
accuracy.set_ylabel("Classification accuracy")
timescale.set_ylabel("Training time in seconds")
accuracy.legend(loc='best')
timescale.legend(loc='best')
# 可視化決策麵,向下投影到數據集的前兩個主要組成部分
pca = PCA(n_components=8).fit(data_train)
X = pca.transform(data_train)
# Generate grid along first two principal components
multiples = np.arange(-2, 2, 0.1)
# steps along first component
first = multiples[:, np.newaxis] * pca.components_[0, :]
# steps along second component
second = multiples[:, np.newaxis] * pca.components_[1, :]
# combine
grid = first[np.newaxis, :, :] + second[:, np.newaxis, :]
flat_grid = grid.reshape(-1, data.shape[1])
# 圖像標題
titles = ['SVC with rbf kernel',
'SVC (linear kernel)\n with Fourier rbf feature map\n'
'n_components=100',
'SVC (linear kernel)\n with Nystroem rbf feature map\n'
'n_components=100']
plt.tight_layout()
plt.figure(figsize=(12, 5))
# 預測和繪圖
for i, clf in enumerate((kernel_svm, nystroem_approx_svm,
fourier_approx_svm)):
# Plot the decision boundary. For that, we will assign a color to each
# point in the mesh [x_min, x_max]x[y_min, y_max].
plt.subplot(1, 3, i + 1)
Z = clf.predict(flat_grid)
# Put the result into a color plot
Z = Z.reshape(grid.shape[:-1])
plt.contourf(multiples, multiples, Z, cmap=plt.cm.Paired)
plt.axis('off')
# Plot also the training points
plt.scatter(X[:, 0], X[:, 1], c=targets_train, cmap=plt.cm.Paired,
edgecolors=(0, 0, 0))
plt.title(titles[i])
plt.tight_layout()
plt.show()
代碼執行
代碼運行時間大約:0分2.188秒。
運行代碼輸出的圖片內容如下:
源碼下載
- Python版源碼文件: plot_kernel_approximation.py
- Jupyter Notebook版源碼文件: plot_kernel_approximation.ipynb
