scream() 確保 data 的結構與原型 ptype 相同。在底層,使用 vctrs::vec_cast() ,它將 data 的每一列轉換為與 ptype 中相應列相同的類型。
該鑄造強製執行許多重要的結構檢查,包括但不限於:
-
數據類 - 檢查
data中每列的類是否與ptype中相應列的類相同。 -
新級別 - 檢查
data中的因子列與ptype列相比是否沒有任何新級別。如果有新級別,則會發出警告,並將它們強製為NA。此檢查是可選的,可以使用allow_novel_levels = TRUE關閉。 -
級別恢複 - 檢查
data中的因子列與ptype列相比是否缺少任何因子級別。如果存在缺失的關卡,則會將其恢複。
參數
- data
-
包含要檢查其結構的新數據的 DataFrame 。
- ptype
-
將
data轉換為的 DataFrame 原型。這通常是訓練集的 0 行切片。 - allow_novel_levels
-
是否應該允許
data中的新因子水平?最安全的方法是默認方法,當發現新級別時會發出警告,並將它們強製為NA值。將此參數設置為TRUE將忽略所有新級別。這個論點不適用於有序因子。有序因子中不允許出現新穎的級別,因為級別排序是該類型的關鍵部分。
細節
scream() 由forge() 在shrink() 之後但在實際處理完成之前調用。一般來說,你不需要直接調用scream(),因為forge()會為你做這件事。
如果 scream() 用作獨立函數,則最好在其之前調用 shrink(),因為 scream() 中沒有檢查來確保 data 中實際存在所有必需的列名稱。這些檢查存在於 shrink() 中。
因子水平
scream() 嘗試通過恢複缺失的因子水平並警告新水平來提供幫助。下圖概述了從 data 中的列強製到 ptype 中的列時 scream() 如何處理因子級別。
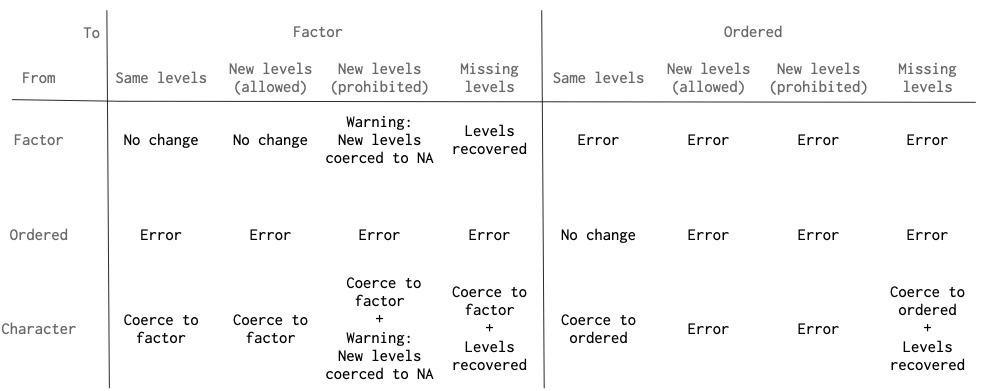
請注意,有序因子處理比因子處理嚴格得多。 data 中的有序因子必須與 ptype 中的有序因子具有完全相同的級別。
例子
# ---------------------------------------------------------------------------
# Setup
train <- iris[1:100, ]
test <- iris[101:150, ]
# mold() is run at model fit time
# and a formula preprocessing blueprint is recorded
x <- mold(log(Sepal.Width) ~ Species, train)
# Inside the result of mold() are the prototype tibbles
# for the predictors and the outcomes
ptype_pred <- x$blueprint$ptypes$predictors
ptype_out <- x$blueprint$ptypes$outcomes
# ---------------------------------------------------------------------------
# shrink() / scream()
# Pass the test data, along with a prototype, to
# shrink() to extract the prototype columns
test_shrunk <- shrink(test, ptype_pred)
# Now pass that to scream() to perform validation checks
# If no warnings / errors are thrown, the checks were
# successful!
scream(test_shrunk, ptype_pred)
#> # A tibble: 50 × 1
#> Species
#> <fct>
#> 1 virginica
#> 2 virginica
#> 3 virginica
#> 4 virginica
#> 5 virginica
#> 6 virginica
#> 7 virginica
#> 8 virginica
#> 9 virginica
#> 10 virginica
#> # ℹ 40 more rows
# ---------------------------------------------------------------------------
# Outcomes
# To also extract the outcomes, use the outcome prototype
test_outcome <- shrink(test, ptype_out)
scream(test_outcome, ptype_out)
#> # A tibble: 50 × 1
#> Sepal.Width
#> <dbl>
#> 1 3.3
#> 2 2.7
#> 3 3
#> 4 2.9
#> 5 3
#> 6 3
#> 7 2.5
#> 8 2.9
#> 9 2.5
#> 10 3.6
#> # ℹ 40 more rows
# ---------------------------------------------------------------------------
# Casting
# scream() uses vctrs::vec_cast() to intelligently convert
# new data to the prototype automatically. This means
# it can automatically perform certain conversions, like
# coercing character columns to factors.
test2 <- test
test2$Species <- as.character(test2$Species)
test2_shrunk <- shrink(test2, ptype_pred)
scream(test2_shrunk, ptype_pred)
#> # A tibble: 50 × 1
#> Species
#> <fct>
#> 1 virginica
#> 2 virginica
#> 3 virginica
#> 4 virginica
#> 5 virginica
#> 6 virginica
#> 7 virginica
#> 8 virginica
#> 9 virginica
#> 10 virginica
#> # ℹ 40 more rows
# It can also recover missing factor levels.
# For example, it is plausible that the test data only had the
# "virginica" level
test3 <- test
test3$Species <- factor(test3$Species, levels = "virginica")
test3_shrunk <- shrink(test3, ptype_pred)
test3_fixed <- scream(test3_shrunk, ptype_pred)
# scream() recovered the missing levels
levels(test3_fixed$Species)
#> [1] "setosa" "versicolor" "virginica"
# ---------------------------------------------------------------------------
# Novel levels
# When novel levels with any data are present in `data`, the default
# is to coerce them to `NA` values with a warning.
test4 <- test
test4$Species <- as.character(test4$Species)
test4$Species[1] <- "new_level"
test4$Species <- factor(
test4$Species,
levels = c(levels(test$Species), "new_level")
)
test4 <- shrink(test4, ptype_pred)
# Warning is thrown
test4_removed <- scream(test4, ptype_pred)
#> Warning: Novel levels found in column 'Species': 'new_level'. The levels have been removed, and values have been coerced to 'NA'.
# Novel level is removed
levels(test4_removed$Species)
#> [1] "setosa" "versicolor" "virginica"
# No warning is thrown
test4_kept <- scream(test4, ptype_pred, allow_novel_levels = TRUE)
# Novel level is kept
levels(test4_kept$Species)
#> [1] "setosa" "versicolor" "virginica" "new_level"
相關用法
- R hardhat standardize 標準化結果
- R hardhat shrink 僅對所需列進行子集化
- R hardhat spruce-multiple 完善多結果預測
- R hardhat validate_prediction_size 確保預測具有正確的行數
- R hardhat default_recipe_blueprint 默認配方藍圖
- R hardhat is_blueprint x 是預處理藍圖嗎?
- R hardhat validate_column_names 確保數據包含所需的列名
- R hardhat default_formula_blueprint 默認公式藍圖
- R hardhat update_blueprint 更新預處理藍圖
- R hardhat weighted_table 加權表
- R hardhat validate_outcomes_are_univariate 確保結果是單變量
- R hardhat get_levels 從 DataFrame 中提取因子水平
- R hardhat add_intercept_column 向數據添加截距列
- R hardhat is_frequency_weights x 是頻率權重向量嗎?
- R hardhat model_offset 提取模型偏移
- R hardhat model_matrix 構建設計矩陣
- R hardhat is_importance_weights x 是重要性權重向量嗎?
- R hardhat run-mold 根據藍圖 Mold()
- R hardhat get_data_classes 從 DataFrame 或矩陣中提取數據類
- R hardhat fct_encode_one_hot 將一個因子編碼為 one-hot 指標矩陣
- R hardhat new_frequency_weights 構建頻率權重向量
- R hardhat validate_no_formula_duplication 確保公式中不出現重複項
- R hardhat default_xy_blueprint 默認 XY 藍圖
- R hardhat validate_outcomes_are_numeric 確保結果都是數字
- R hardhat frequency_weights 頻率權重
注:本文由純淨天空篩選整理自Davis Vaughan等大神的英文原創作品 ? Scream.。非經特殊聲明,原始代碼版權歸原作者所有,本譯文未經允許或授權,請勿轉載或複製。
